3.5. 演習:胸部 X 線画像診断#
深層学習を活用した胸部 X 線画像診断支援システムは、医療分野で注目されている技術です。この技術は、X 線画像を解析し、肺炎や肺がんなどの病変を検出することで、診断をサポートすることを目的としています。医師の診断を補助することで、小さな病変を見逃すリスクを減らし、診断の迅速化や医師の業務負担の軽減に貢献できると期待されています。本節では、深層学習を使って健康者と肺炎患者の胸部 X 線画像を解析し、肺炎の有無を診断するプログラムを作成する方法を学びます。
3.5.1. 演習準備#
3.5.1.1. ライブラリ#
本節で利用するライブラリを読み込みます。ライブラリの読み込み時に ImportError や ModuleNotFoundError が発生した場合は、該当するライブラリをインストールしてください。pytorch_grad_cam を読み込むときに ModuleNotFoundError が発生した場合は、grad-cam パッケージをインストールしてください。
# image
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import PIL.Image
# machine learning
import sklearn.metrics
import torch
import torchvision
# grad-CAM visualization
import cv2
import pytorch_grad_cam
print(f'torch v{torch.__version__}; torchvision v{torchvision.__version__}')
torch v2.8.0+cu126; torchvision v0.23.0+cu126
3.5.1.2. データセット#
本節では、Kermany らの論文[1]で公開されている胸部 X 線画像データセットを利用します。このデータセットには、健康者(NORMAL)と肺炎患者(PNEUMONIA)の胸部 X 線画像が含まれています。また、肺炎患者の画像は、さらに細菌性肺炎とウイルス性肺炎の 2 種類に分類されています(Fig. 3.13)。
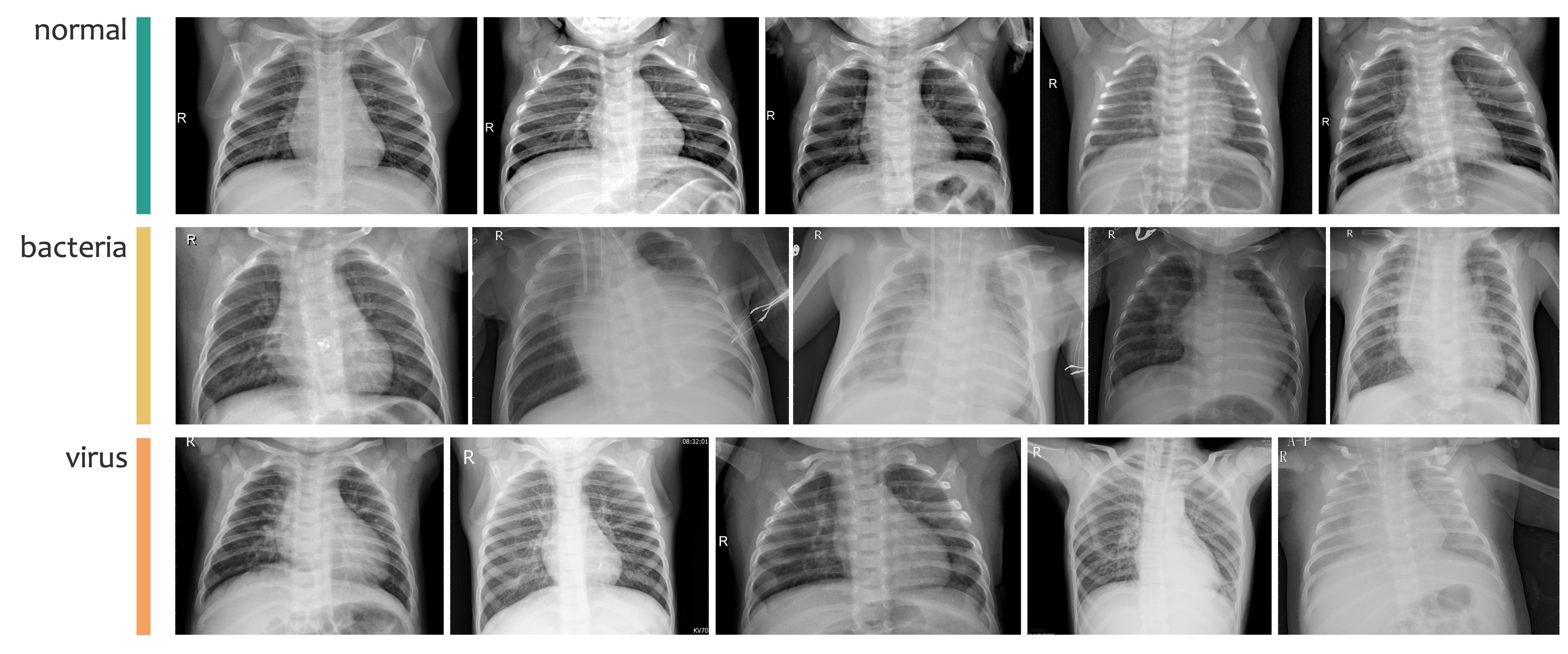
Fig. 3.13 Kermany らのデータセットに含まれる健康者(normal)、細菌性肺炎患者(bacteria)、ウイルス性肺炎患者(virus)の胸部 X 線画像の例。#
このデータセットは、CC-BY 4.0 ライセンスのもとで Mendeley Data に公開されています。このライセンスにより、著作権表示を行うことで、編集や再配布を含む自由な利用が可能です。
本節では、オリジナルのデータセットからランダムに抽出した 360 枚の画像を使用して作成した小規模なデータセットを利用します。このデータセットは、訓練サブセット、検証サブセット、テストサブセットの 3 つに分かれています。訓練サブセットには各カテゴリ(normal、bacteria、virus)の画像がそれぞれ 100 枚含まれており、検証サブセットとテストサブセットには、それぞれ各カテゴリ 10 枚の画像が含まれています。
本節で利用するデータセットは、Jupyter Notebook 上では、次のコマンドを実行することでダウンロードできます。
!wget https://dl.biopapyrus.jp/data/chestxray_pneumonia.zip
!unzip chestxray_pneumonia.zip
3.5.1.3. 画像前処理#
畳み込みニューラルネットワークは、ニューロンの数などが固定されているため、入力する画像のサイズにも制限があります。例えば、本節で使用する EfficientNet V2(efficientnet_v2_m) [2] では、480×480 ピクセルの正方形画像を入力として設計されています。また、PyTorch ではすべてのデータをテンソル形式で扱う必要があります。そのため、畳み込みニューラルネットワークに画像を入力する前に、画像サイズを適切に調整し、テンソル型に変換するといった前処理を行う必要があります。以下では、この前処理の手順を定義します。
class SquareResize:
def __init__(self, shape=480, bg_color = (0, 0, 0)):
self.shape = shape
self.bg_color = tuple(bg_color)
def __call__(self, img):
w, h = img.size
img_square = None
if w == h:
img_square = img
elif w > h:
img_square = PIL.Image.new(img.mode, (w, w), self.bg_color)
img_square.paste(img, (0, (w - h) // 2))
else:
img_square = PIL.Image.new(img.mode, (h, h), self.bg_color)
img_square.paste(img, ((h - w) // 2, 0))
img_square = img_square.resize((self.shape, self.shape))
return img_square
transform = torchvision.transforms.Compose([
SquareResize(),
torchvision.transforms.ToTensor(),
torchvision.transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
3.5.1.4. 計算デバイス#
計算を行うデバイスを設定します。PyTorch が GPU を認識できる場合は GPU を利用し、認識できない場合は CPU を使用するように設定します。
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
3.5.2. モデル構築#
EfficientNet V2 のアーキテクチャを利用してモデルを構築します。torchvision.models モジュールで提供されている EfficientNet V2 には、efficientnet_v2_s、efficientnet_v2_m、efficientnet_v2_l の 3 つのサブタイプがあります。それぞれ、入力画像のサイズが異なり、入力画像サイズが大きいほど性能が向上しますが、モデルのパラメータが多くなり、訓練時間も長くなります。本節では、efficientnet_v2_m を利用します。このアーキテクチャは、480×480 ピクセルの画像を入力として設計されています。
torchvision.models モジュールで提供されているアーキテクチャは、飛行機や車、人など、1000 種類の一般的な物体を分類するように設計されています。これに対して、本節では、normal、bacteria、virus の 3 カテゴリの分類問題を扱います。そのため、torchvision.models モジュールから読み込んだ EfficientNet V2 の出力層のユニット数を 3 に変更する必要があります。この修正作業はモデルを呼び出すたびに行う必要があり、手間がかかります。そこで、一連の処理を関数化してから利用します。
def efficientnet_v2(num_classes, weights=None):
model = torchvision.models.efficientnet_v2_m(weights='DEFAULT')
in_features = model.classifier[1].in_features
model.classifier[1] = torch.nn.Linear(in_features, num_classes)
if weights is not None:
model.load_state_dict(torch.load(weights))
return model
model = efficientnet_v2(3)
3.5.3. モデル訓練#
モデルが学習データを効率よく学習できるようにするため、損失関数(criterion)、学習アルゴリズム(optimizer)、学習率(lr)、および学習率を調整するスケジューラ(lr_scheduler)を設定します。
criterion = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.00003)
lr_scheduler = torch.optim.lr_scheduler.StepLR(optimizer, step_size=3, gamma=0.1)
次に、訓練データと検証データを読み込み、モデルが入力できる形式に整えます。
train_dataset = torchvision.datasets.ImageFolder('chestxray_pneumonia/train', transform=transform)
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=8, shuffle=True)
valid_dataset = torchvision.datasets.ImageFolder('chestxray_pneumonia/valid', transform=transform)
valid_loader = torch.utils.data.DataLoader(valid_dataset, batch_size=8, shuffle=False)
準備が整ったら、訓練を開始します。訓練プロセスでは、訓練と検証を交互に繰り返します。訓練では、訓練データを使ってモデルのパラメータを更新し、その際の損失(誤差)を記録します。検証では、検証データを使ってモデルの予測性能(正解率)を計算し、その結果を記録します。このサイクルを繰り返すことで、モデルの精度を少しずつ向上させていきます。
model.to(device)
num_epochs = 15
metric_dict = []
for epoch in range(num_epochs):
# training phase
model.train()
running_loss = 0.0
n_correct_train = 0
n_train_samples = 0
for images, labels in train_loader:
images = images.to(device)
labels = labels.to(device)
outputs = model(images)
loss = criterion(outputs, labels)
_, predicted_labels = torch.max(outputs.data, 1)
n_correct_train += torch.sum(predicted_labels == labels).item()
n_train_samples += labels.size(0)
running_loss += loss.item() / len(train_loader)
optimizer.zero_grad()
loss.backward()
optimizer.step()
lr_scheduler.step()
# validation phase
model.eval()
n_correct_valid = 0
n_valid_samples = 0
with torch.no_grad():
for images, labels in valid_loader:
images = images.to(device)
labels = labels.to(device)
outputs = model(images)
_, predicted_labels = torch.max(outputs.data, 1)
n_correct_valid += torch.sum(predicted_labels == labels).item()
n_valid_samples += labels.size(0)
metric_dict.append({
'epoch': epoch + 1,
'train_loss': running_loss,
'train_acc': n_correct_train / n_train_samples,
'valid_acc': n_correct_valid / n_valid_samples
})
print(metric_dict[-1])
Show code cell output
{'epoch': 1, 'train_loss': 0.9793290417445331, 'train_acc': 0.5466666666666666, 'valid_acc': 0.6333333333333333}
{'epoch': 2, 'train_loss': 0.6822633217824132, 'train_acc': 0.72, 'valid_acc': 0.8666666666666667}
{'epoch': 3, 'train_loss': 0.47430643282438584, 'train_acc': 0.8366666666666667, 'valid_acc': 0.9}
{'epoch': 4, 'train_loss': 0.3470618826778312, 'train_acc': 0.8733333333333333, 'valid_acc': 0.9}
{'epoch': 5, 'train_loss': 0.28201773939164054, 'train_acc': 0.9166666666666666, 'valid_acc': 0.9}
{'epoch': 6, 'train_loss': 0.2683706722761454, 'train_acc': 0.94, 'valid_acc': 0.9}
{'epoch': 7, 'train_loss': 0.2694516115282712, 'train_acc': 0.9266666666666666, 'valid_acc': 0.8666666666666667}
{'epoch': 8, 'train_loss': 0.28514728734367767, 'train_acc': 0.9066666666666666, 'valid_acc': 0.8666666666666667}
{'epoch': 9, 'train_loss': 0.2539483091156734, 'train_acc': 0.9333333333333333, 'valid_acc': 0.9}
{'epoch': 10, 'train_loss': 0.2632817664232694, 'train_acc': 0.9333333333333333, 'valid_acc': 0.8666666666666667}
{'epoch': 11, 'train_loss': 0.22924590346060297, 'train_acc': 0.9433333333333334, 'valid_acc': 0.8666666666666667}
{'epoch': 12, 'train_loss': 0.2477570366310446, 'train_acc': 0.93, 'valid_acc': 0.9}
{'epoch': 13, 'train_loss': 0.24513497889826175, 'train_acc': 0.93, 'valid_acc': 0.8666666666666667}
{'epoch': 14, 'train_loss': 0.2552205533965638, 'train_acc': 0.92, 'valid_acc': 0.9}
{'epoch': 15, 'train_loss': 0.24042309859865593, 'train_acc': 0.9466666666666667, 'valid_acc': 0.9}
訓練データに対する損失と検証データに対する正解率を可視化し、訓練過程を評価します。
Show code cell source
metric_dict = pd.DataFrame(metric_dict)
fig, ax = plt.subplots(1, 2)
ax[0].plot(metric_dict['epoch'], metric_dict['train_loss'])
ax[0].set_xlabel('epoch')
ax[0].set_ylabel('loss')
ax[0].set_title('Train')
ax[1].plot(metric_dict['epoch'], metric_dict['valid_acc'])
ax[1].set_ylim(0, 1)
ax[1].set_xlabel('epoch')
ax[1].set_ylabel('accuracy')
ax[1].set_title('Validation')
plt.tight_layout()
fig.show()
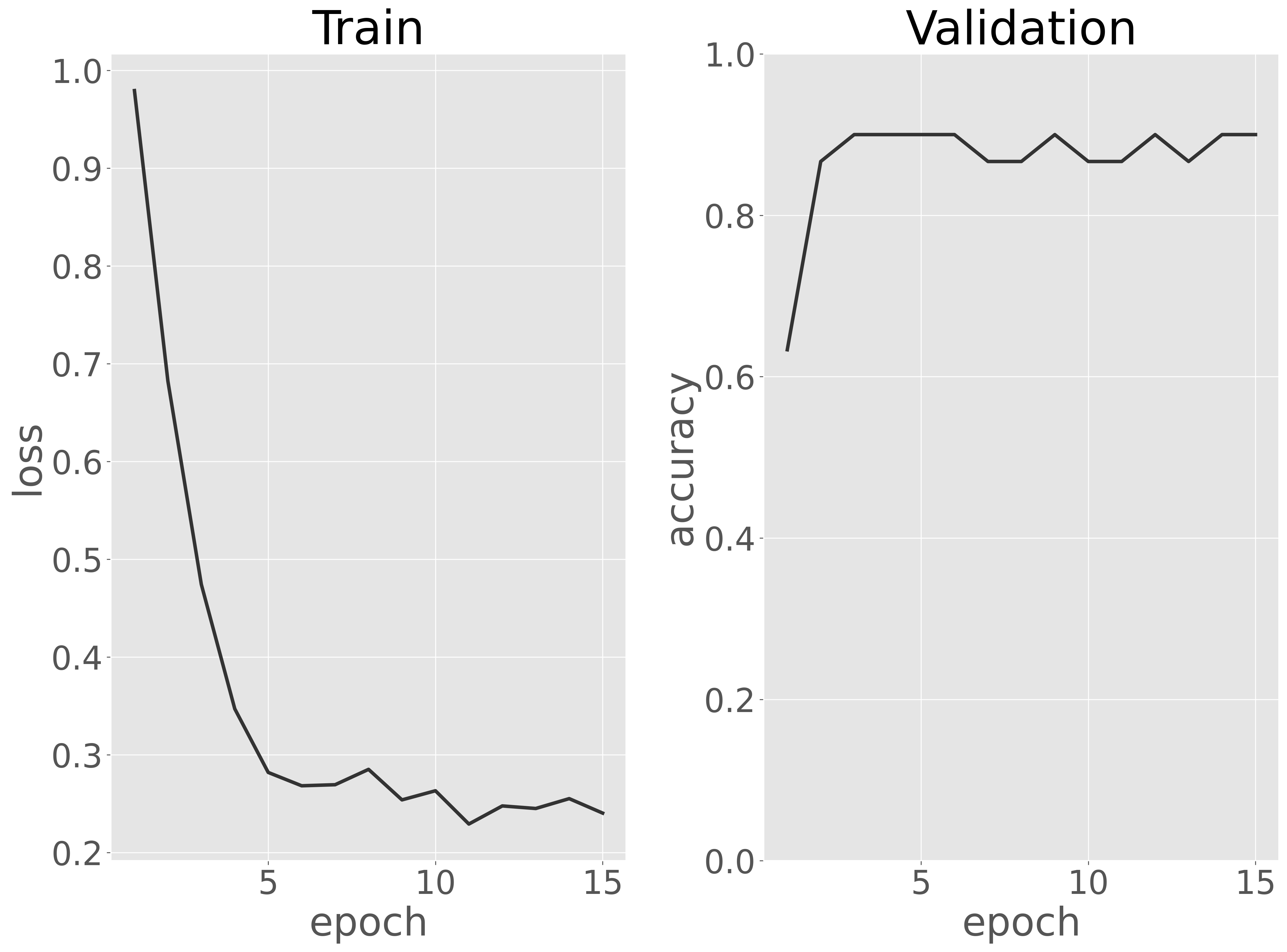
可視化の結果、エポックが増えるにつれて訓練データに対する損失が減少し、10 エポック前後で収束し始めたことが確認できました。また、検証データに対する分類性能(正解率)は、最初の数エポックからほぼ最大値に達しており、EfficientNet V2 は数エポックの学習だけで十分であることがわかります。
次に、同じ手順を異なる深層ニューラルネットワークアーキテクチャ(DenseNet や Inception など)に対して実施し、それぞれのアーキテクチャの検証性能を比較します。そして、このデータセットに最適なアーキテクチャを選定します。ただし、本節ではモデル(アーキテクチャ)選択を行わずに、EfficientNet V2 を最適なアーキテクチャとして採用し、次のステップに進みます。
次のステップでは、訓練サブセットと検証サブセットを統合し、最適なアーキテクチャを最初から訓練します。
!mkdir chestxray_pneumonia/trainvalid
!cp -r chestxray_pneumonia/train/* chestxray_pneumonia/trainvalid
!cp -r chestxray_pneumonia/valid/* chestxray_pneumonia/trainvalid
最適なモデルを選択する段階で、数エポックの訓練だけでも十分に高い予測性能を獲得できたことがわかったので、ここでは訓練サブセットと検証サブセットを統合したデータに対して 5 エポックだけ訓練させます。
# model
model = efficientnet_v2(3)
model.to(device)
# training params
criterion = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.00003)
lr_scheduler = torch.optim.lr_scheduler.StepLR(optimizer, step_size=3, gamma=0.1)
# training data
train_dataset = torchvision.datasets.ImageFolder('chestxray_pneumonia/trainvalid', transform=transform)
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=8, shuffle=True)
# training
num_epochs = 5
metric_dict = []
for epoch in range(num_epochs):
model.train()
running_loss = 0.0
n_correct_train = 0
n_train_samples = 0
for images, labels in train_loader:
images = images.to(device)
labels = labels.to(device)
outputs = model(images)
loss = criterion(outputs, labels)
_, predicted_labels = torch.max(outputs.data, 1)
n_correct_train += torch.sum(predicted_labels == labels).item()
n_train_samples += labels.size(0)
running_loss += loss.item() / len(train_loader)
optimizer.zero_grad()
loss.backward()
optimizer.step()
lr_scheduler.step()
metric_dict.append({
'epoch': epoch + 1,
'train_loss': running_loss,
'train_acc': n_correct_train / n_train_samples,
})
print(metric_dict[-1])
Show code cell output
{'epoch': 1, 'train_loss': 0.9539776912757328, 'train_acc': 0.6}
{'epoch': 2, 'train_loss': 0.5493250836928686, 'train_acc': 0.803030303030303}
{'epoch': 3, 'train_loss': 0.3373820873953047, 'train_acc': 0.8878787878787879}
{'epoch': 4, 'train_loss': 0.2674782662874176, 'train_acc': 0.9212121212121213}
{'epoch': 5, 'train_loss': 0.1840237839413541, 'train_acc': 0.9333333333333333}
訓練が完了したら、訓練済みモデルの重みをファイルに保存します。
model = model.to('cpu')
torch.save(model.state_dict(), 'chestxray_pneumonia.pth')
3.5.4. モデル評価#
最適なモデルが得られたら、次にテストデータを用いてモデルを詳細に評価します。正解率だけでなく、適合率、再現率、F1 スコアなどの評価指標を計算し、モデルを総合的に評価します。まず、テストデータをモデルに入力し、その予測結果を取得します。
test_dataset = torchvision.datasets.ImageFolder('chestxray_pneumonia/test', transform=transform)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size=8, shuffle=False)
model = efficientnet_v2(3, 'chestxray_pneumonia.pth')
model.to(device)
model.eval()
pred_labels = []
true_labels = []
with torch.no_grad():
for images, labels in test_loader:
images = images.to(device)
labels = labels.to(device)
outputs = model(images)
_, _labels = torch.max(outputs.data, 1)
#print(_labels)
pred_labels.extend(_labels.cpu().detach().numpy().tolist())
true_labels.extend(labels.cpu().detach().numpy().tolist())
pred_labels = [test_dataset.classes[_] for _ in pred_labels]
true_labels = [test_dataset.classes[_] for _ in true_labels]
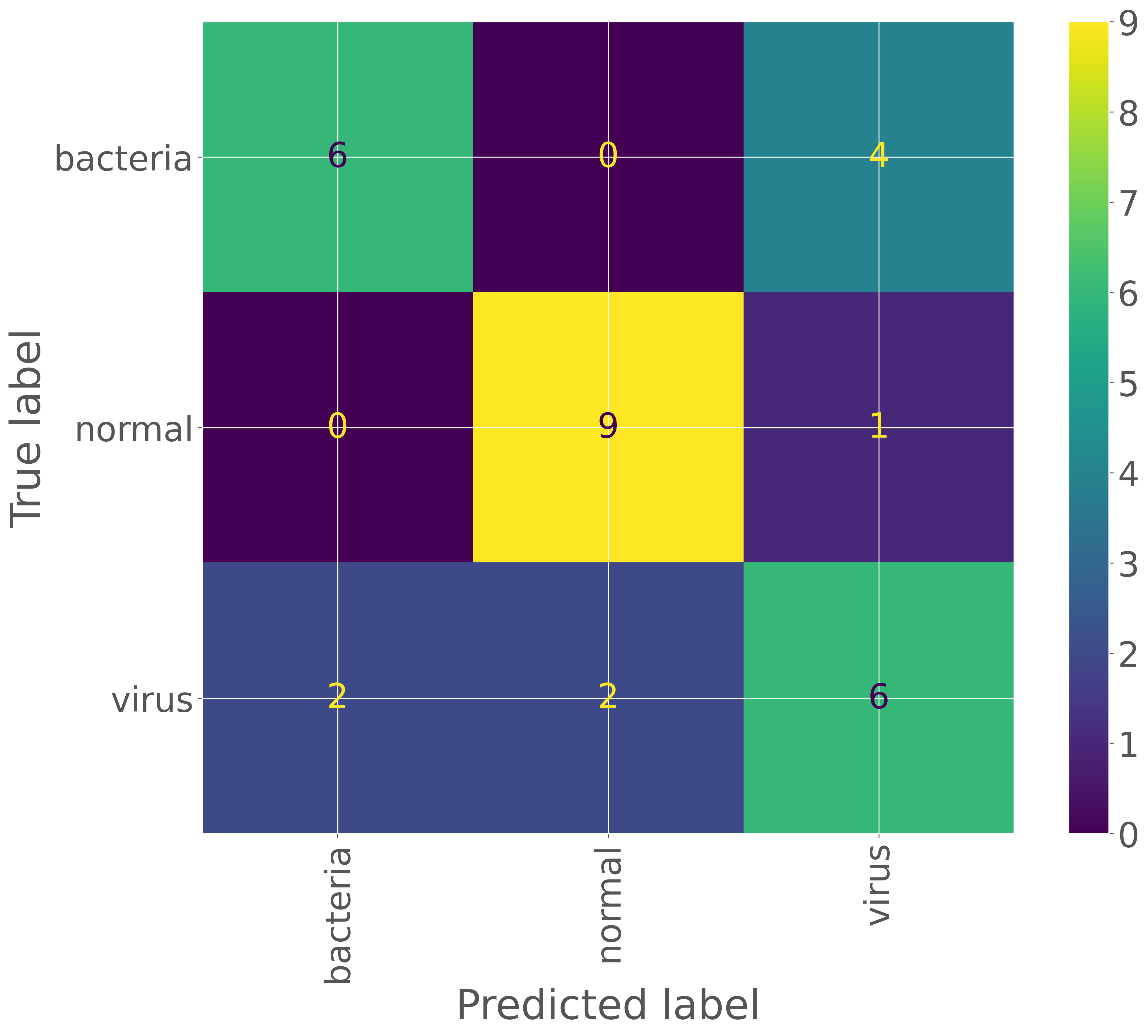
次に、予測結果とラベルを比較し、混同行列を作成します。これにより、間違いやすいカテゴリを特定することができます。
cm = sklearn.metrics.confusion_matrix(true_labels, pred_labels)
print(cm)
[[6 0 4]
[0 9 1]
[2 2 6]]
cmp = sklearn.metrics.ConfusionMatrixDisplay(cm, display_labels=test_dataset.classes)
cmp.plot(xticks_rotation='vertical')
<sklearn.metrics._plot.confusion_matrix.ConfusionMatrixDisplay at 0x7fc978ab6bd0>
それぞれのクラスに対する適合率、再現率、F1 スコアなどは、scikit-learn ライブラリを利用して計算します。
pd.DataFrame(sklearn.metrics.classification_report(true_labels, pred_labels, output_dict=True))
| bacteria | normal | virus | accuracy | macro avg | weighted avg | |
|---|---|---|---|---|---|---|
| precision | 0.750000 | 0.818182 | 0.545455 | 0.7 | 0.704545 | 0.704545 |
| recall | 0.600000 | 0.900000 | 0.600000 | 0.7 | 0.700000 | 0.700000 |
| f1-score | 0.666667 | 0.857143 | 0.571429 | 0.7 | 0.698413 | 0.698413 |
| support | 10.000000 | 10.000000 | 10.000000 | 0.7 | 30.000000 | 30.000000 |
3.5.5. 推論#
推論を行う際には、訓練や評価時と同様に、torchvision.models モジュールから EfficientNet V2 のアーキテクチャを読み込み、出力層のクラス数を設定します。その後、load_state_dict メソッドを使用して訓練済みの重みファイルをモデルにロードします。これらの処理はすでに関数化(efficientnet_v2)されているため、その関数を利用して簡単に実行できます。
labels = ['bacteria', 'normal', 'virus']
model = efficientnet_v2(3, 'chestxray_pneumonia.pth')
model.to(device)
model.eval()
Show code cell output
EfficientNet(
(features): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(3, 24, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(1): BatchNorm2d(24, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Sequential(
(0): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(24, 24, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(24, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
)
(stochastic_depth): StochasticDepth(p=0.0, mode=row)
)
(1): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(24, 24, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(24, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
)
(stochastic_depth): StochasticDepth(p=0.0035087719298245615, mode=row)
)
(2): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(24, 24, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(24, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
)
(stochastic_depth): StochasticDepth(p=0.007017543859649123, mode=row)
)
)
(2): Sequential(
(0): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(24, 96, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(1): BatchNorm2d(96, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(96, 48, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(48, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.010526315789473686, mode=row)
)
(1): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(48, 192, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(192, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(192, 48, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(48, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.014035087719298246, mode=row)
)
(2): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(48, 192, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(192, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(192, 48, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(48, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.017543859649122806, mode=row)
)
(3): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(48, 192, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(192, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(192, 48, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(48, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.02105263157894737, mode=row)
)
(4): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(48, 192, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(192, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(192, 48, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(48, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.024561403508771933, mode=row)
)
)
(3): Sequential(
(0): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(48, 192, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(1): BatchNorm2d(192, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(192, 80, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(80, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.028070175438596492, mode=row)
)
(1): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(80, 320, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(320, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(320, 80, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(80, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.031578947368421054, mode=row)
)
(2): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(80, 320, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(320, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(320, 80, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(80, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.03508771929824561, mode=row)
)
(3): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(80, 320, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(320, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(320, 80, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(80, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.03859649122807018, mode=row)
)
(4): FusedMBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(80, 320, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): BatchNorm2d(320, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(320, 80, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(80, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.04210526315789474, mode=row)
)
)
(4): Sequential(
(0): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(80, 320, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(320, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(320, 320, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), groups=320, bias=False)
(1): BatchNorm2d(320, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(320, 20, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(20, 320, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(320, 160, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(160, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.0456140350877193, mode=row)
)
(1): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(160, 640, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(640, 640, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=640, bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(640, 40, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(40, 640, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(640, 160, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(160, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.04912280701754387, mode=row)
)
(2): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(160, 640, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(640, 640, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=640, bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(640, 40, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(40, 640, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(640, 160, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(160, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.05263157894736842, mode=row)
)
(3): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(160, 640, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(640, 640, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=640, bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(640, 40, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(40, 640, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(640, 160, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(160, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.056140350877192984, mode=row)
)
(4): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(160, 640, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(640, 640, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=640, bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(640, 40, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(40, 640, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(640, 160, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(160, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.05964912280701755, mode=row)
)
(5): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(160, 640, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(640, 640, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=640, bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(640, 40, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(40, 640, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(640, 160, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(160, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.06315789473684211, mode=row)
)
(6): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(160, 640, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(640, 640, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=640, bias=False)
(1): BatchNorm2d(640, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(640, 40, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(40, 640, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(640, 160, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(160, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.06666666666666667, mode=row)
)
)
(5): Sequential(
(0): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(160, 960, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(960, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(960, 960, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=960, bias=False)
(1): BatchNorm2d(960, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(960, 40, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(40, 960, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(960, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.07017543859649122, mode=row)
)
(1): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.0736842105263158, mode=row)
)
(2): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.07719298245614035, mode=row)
)
(3): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.08070175438596493, mode=row)
)
(4): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.08421052631578949, mode=row)
)
(5): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.08771929824561403, mode=row)
)
(6): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.0912280701754386, mode=row)
)
(7): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.09473684210526316, mode=row)
)
(8): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.09824561403508773, mode=row)
)
(9): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.10175438596491229, mode=row)
)
(10): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.10526315789473684, mode=row)
)
(11): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.10877192982456141, mode=row)
)
(12): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.11228070175438597, mode=row)
)
(13): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 176, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(176, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.11578947368421054, mode=row)
)
)
(6): Sequential(
(0): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(176, 1056, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1056, 1056, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), groups=1056, bias=False)
(1): BatchNorm2d(1056, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1056, 44, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(44, 1056, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1056, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.1192982456140351, mode=row)
)
(1): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.12280701754385964, mode=row)
)
(2): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.12631578947368421, mode=row)
)
(3): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.1298245614035088, mode=row)
)
(4): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.13333333333333333, mode=row)
)
(5): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.1368421052631579, mode=row)
)
(6): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.14035087719298245, mode=row)
)
(7): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.14385964912280705, mode=row)
)
(8): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.1473684210526316, mode=row)
)
(9): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.15087719298245614, mode=row)
)
(10): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.1543859649122807, mode=row)
)
(11): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.15789473684210525, mode=row)
)
(12): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.16140350877192985, mode=row)
)
(13): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.1649122807017544, mode=row)
)
(14): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.16842105263157897, mode=row)
)
(15): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.17192982456140352, mode=row)
)
(16): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.17543859649122806, mode=row)
)
(17): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 304, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(304, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.17894736842105266, mode=row)
)
)
(7): Sequential(
(0): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(304, 1824, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(1824, 1824, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=1824, bias=False)
(1): BatchNorm2d(1824, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(1824, 76, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(76, 1824, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(1824, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(512, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.1824561403508772, mode=row)
)
(1): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(512, 3072, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(3072, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(3072, 3072, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=3072, bias=False)
(1): BatchNorm2d(3072, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(3072, 128, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(128, 3072, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(3072, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(512, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.18596491228070178, mode=row)
)
(2): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(512, 3072, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(3072, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(3072, 3072, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=3072, bias=False)
(1): BatchNorm2d(3072, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(3072, 128, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(128, 3072, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(3072, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(512, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.18947368421052632, mode=row)
)
(3): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(512, 3072, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(3072, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(3072, 3072, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=3072, bias=False)
(1): BatchNorm2d(3072, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(3072, 128, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(128, 3072, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(3072, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(512, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.19298245614035087, mode=row)
)
(4): MBConv(
(block): Sequential(
(0): Conv2dNormActivation(
(0): Conv2d(512, 3072, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(3072, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(1): Conv2dNormActivation(
(0): Conv2d(3072, 3072, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), groups=3072, bias=False)
(1): BatchNorm2d(3072, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
(2): SqueezeExcitation(
(avgpool): AdaptiveAvgPool2d(output_size=1)
(fc1): Conv2d(3072, 128, kernel_size=(1, 1), stride=(1, 1))
(fc2): Conv2d(128, 3072, kernel_size=(1, 1), stride=(1, 1))
(activation): SiLU(inplace=True)
(scale_activation): Sigmoid()
)
(3): Conv2dNormActivation(
(0): Conv2d(3072, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(512, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
)
)
(stochastic_depth): StochasticDepth(p=0.19649122807017547, mode=row)
)
)
(8): Conv2dNormActivation(
(0): Conv2d(512, 1280, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): BatchNorm2d(1280, eps=0.001, momentum=0.1, affine=True, track_running_stats=True)
(2): SiLU(inplace=True)
)
)
(avgpool): AdaptiveAvgPool2d(output_size=1)
(classifier): Sequential(
(0): Dropout(p=0.3, inplace=True)
(1): Linear(in_features=1280, out_features=3, bias=True)
)
)
このモデルを使用して推論を行います。細菌性肺炎(bacteria)の画像を 1 枚指定し、訓練時と同様の前処理を行います。その後、前処理した画像をモデルに入力し、モデルから予測結果が出力されます。
image_path = 'chestxray_pneumonia/test/bacteria/person5_bacteria_16.jpeg'
image = PIL.Image.open(image_path).convert('RGB')
input_tensor = transform(image).unsqueeze(0).to(device)
with torch.no_grad():
score = model(input_tensor)[0]
output = pd.DataFrame({
'class': labels,
'probability': torch.softmax(score, axis=0).cpu().detach().numpy()
})
output
| class | probability | |
|---|---|---|
| 0 | bacteria | 0.922189 |
| 1 | normal | 0.008202 |
| 2 | virus | 0.069610 |
次に、別の例を見てみましょう。ウィルス性肺炎(virus)の画像をモデルに入力し、推論を行います。
image_path = 'chestxray_pneumonia/test/virus/person111_virus_212.jpeg'
image = PIL.Image.open(image_path).convert('RGB')
input_tensor = transform(image).unsqueeze(0).to(device)
with torch.no_grad():
score = model(input_tensor)[0]
pd.DataFrame({
'class': labels,
'probability': torch.softmax(score, axis=0).cpu().detach().numpy()
})
| class | probability | |
|---|---|---|
| 0 | bacteria | 0.003860 |
| 1 | normal | 0.962097 |
| 2 | virus | 0.034043 |
3.5.6. 分類根拠の可視化#
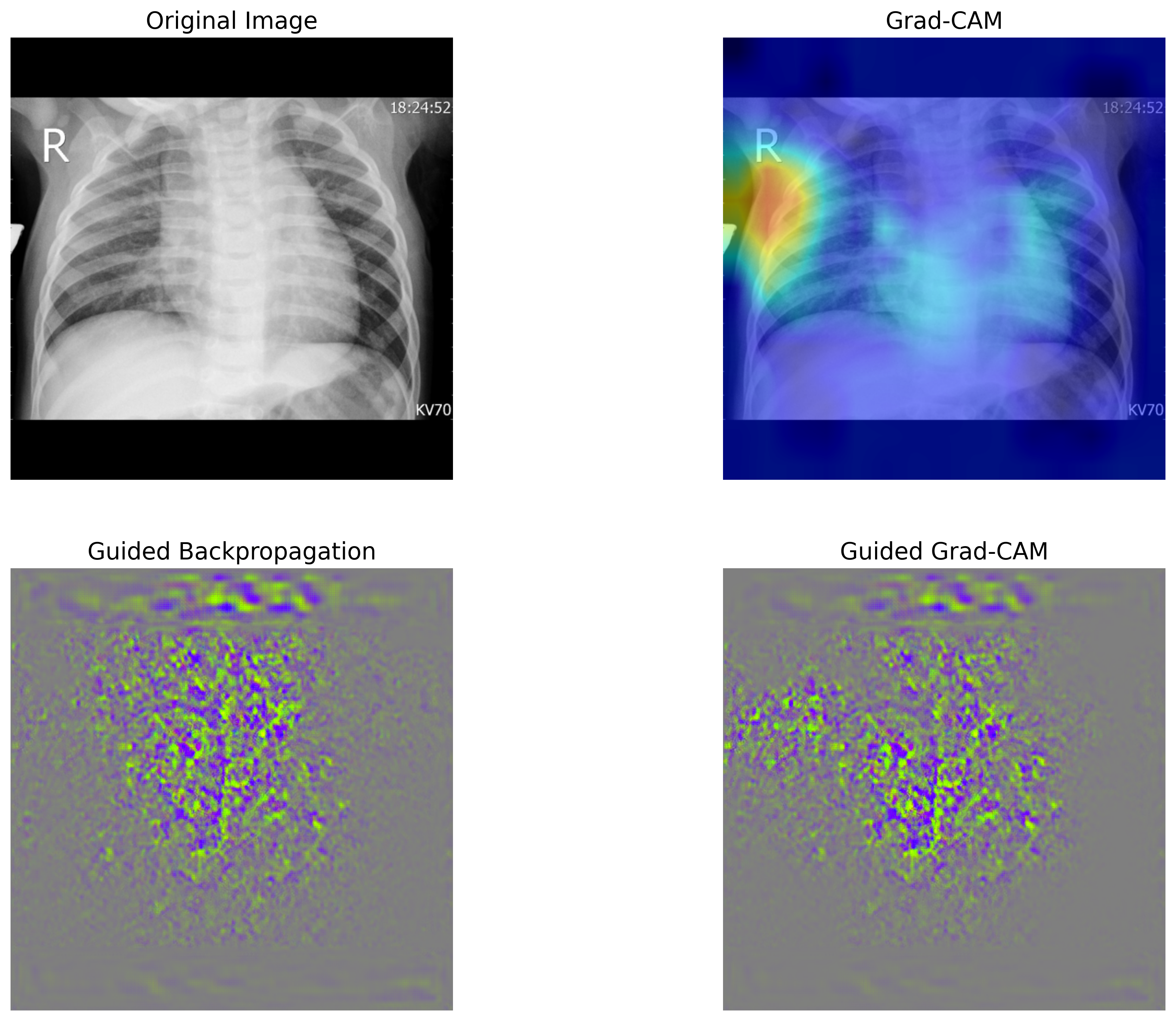
畳み込みニューラルネットワークを用いた画像分類では、畳み込み層で抽出された特徴マップが分類に大きな影響を与えています。そのため、最後の畳み込み層で得られた特徴マップと、それに対応する重みを可視化することで、モデルがどの部分に注目して分類を行ったのか、つまり判断の根拠を明確にすることができます。
本節では、Grad-CAM(Gradient-weighted Class Activation Mapping)および Guided Grad-CAM という手法を用いて、モデルの判断根拠を可視化します。可視化には Python の grad-cam パッケージを使用します。必要に応じてインストールし、grad-cam のチュートリアルを参考にしながら、Grad-CAM および Guided Grad-CAM を計算し、可視化するための関数を定義します。
def viz(image_path):
labels = ['bacteria', 'normal', 'virus']
model = efficientnet_v2(3, 'chestxray_pneumonia.pth')
model.to(device)
model.eval()
# load image
image = PIL.Image.open(image_path).convert('RGB')
input_tensor = transform(image).unsqueeze(0).to(device)
rgb_img = cv2.imread(image_path, 1)[:, :, ::-1]
rgb_img = np.float32(np.array(SquareResize()(image))) / 255
# Grad-CAM
with pytorch_grad_cam.GradCAM(model=model, target_layers=[model.features[8]]) as cam:
cam.batch_size = 8
grayscale_cam = cam(input_tensor=input_tensor, targets=None,aug_smooth=True, eigen_smooth=True)
grayscale_cam = grayscale_cam[0, :]
cam_image = pytorch_grad_cam.utils.image.show_cam_on_image(rgb_img, grayscale_cam, use_rgb=True)
prob = torch.softmax(cam.outputs[0], axis=0).cpu().detach().numpy()
gb_model = pytorch_grad_cam.GuidedBackpropReLUModel(model=model, device=device)
gb = gb_model(input_tensor, target_category=None)
cam_mask = np.stack([grayscale_cam, grayscale_cam, grayscale_cam], axis=-1)
cam_gb = pytorch_grad_cam.utils.image.deprocess_image(cam_mask * gb)
gb = pytorch_grad_cam.utils.image.deprocess_image(gb)
# plot
fig, ax = plt.subplots(2, 2)
ax[0, 0].imshow(rgb_img)
ax[0, 0].axis('off')
ax[0, 0].set_title('Original Image', fontsize=16)
ax[0, 1].imshow(cam_image)
ax[0, 1].axis('off')
ax[0, 1].set_title('Grad-CAM', fontsize=16)
ax[1, 0].imshow(gb)
ax[1, 0].axis('off')
ax[1, 0].set_title('Guided Backpropagation', fontsize=16)
ax[1, 1].imshow(cam_gb)
ax[1, 1].axis('off')
ax[1, 1].set_title('Guided Grad-CAM', fontsize=16)
print(pd.DataFrame({'class': labels, 'probability': prob}))
fig.show()
次に、いくつかの画像をこの可視化関数に入力し、モデルの予測結果とその判断根拠を可視化します。これにより、モデルがどの部分に注目して分類を行ったのかを視覚的に確認することができます。必要に応じて、他の画像を入力し、それぞれの分類結果と判断根拠を可視化してみてください。
viz('chestxray_pneumonia/test/bacteria/person5_bacteria_16.jpeg')
class probability
0 bacteria 0.558184
1 normal 0.185698
2 virus 0.256119
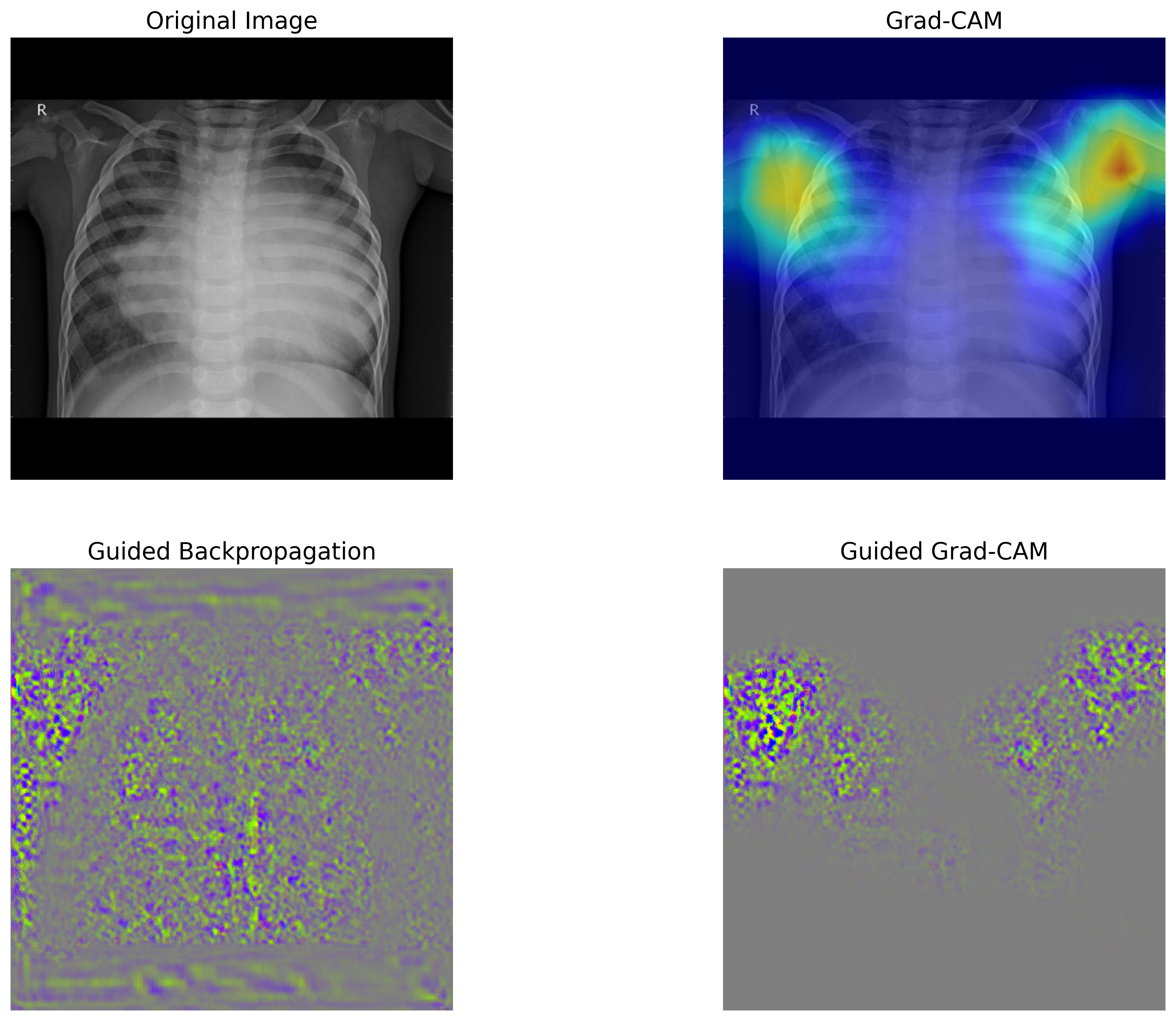
viz('chestxray_pneumonia/test/virus/person1365_virus_2348.jpeg')
class probability
0 bacteria 0.100765
1 normal 0.072158
2 virus 0.827076